Gaussian job submission using g16sub
Last Update: Apr 19, 2024.
Introduction
Aim of this page is to show how to run Gaussian jobs with g16sub command.
- Preparations (transfer input file)
- Submit a Gaussian job with g16sub
- Verify job status (jobinfo)
- Job finished...
- Run formchk
Please check g16sub/g09sub page of the reference manual for g16sub options and tips, too.
Preparations
Following conditions must be satisfied beforehand.
- You can login to RCCS login server (ccfep) with SSH.
- You can send/receive files to/from RCCS via scp or sftp.
- Your Gaussian input file. Just to test the procedure, please use sample input below.
Please visit Quick Start Guide page if ssh, scp, sftp settings are not yet completed.
sample Gaussian input gile
In the following, we use ch3cl.gjf file (.gjf is one of the common file extensions of Gaussian input file) as an example. We need to send this file to RCCS first, and then run Gaussian.
%chk=ch3cl.chk
# HF/6-31G(d,p) Opt
methyl chloride
0,1
C -0.000004 1.127470 0.000000
H -0.511417 1.468491 0.885898
H -0.511417 1.468491 -0.885898
H 1.022922 1.468527 0.000000
Cl -0.000004 -0.657078 0.0000
Please note that %mem, %nprocshared, %cpu are usually overwritten by g16sub/g09sub. Number of CPU cores can be specified by -np option of g16sub/g09sub. For the memory amount, you don't need to set it manually. This is because reasonable value will be automatically assigned according to the jobtype and number of CPU cores.
File Transfer (1)
We employ WinSCP in this example.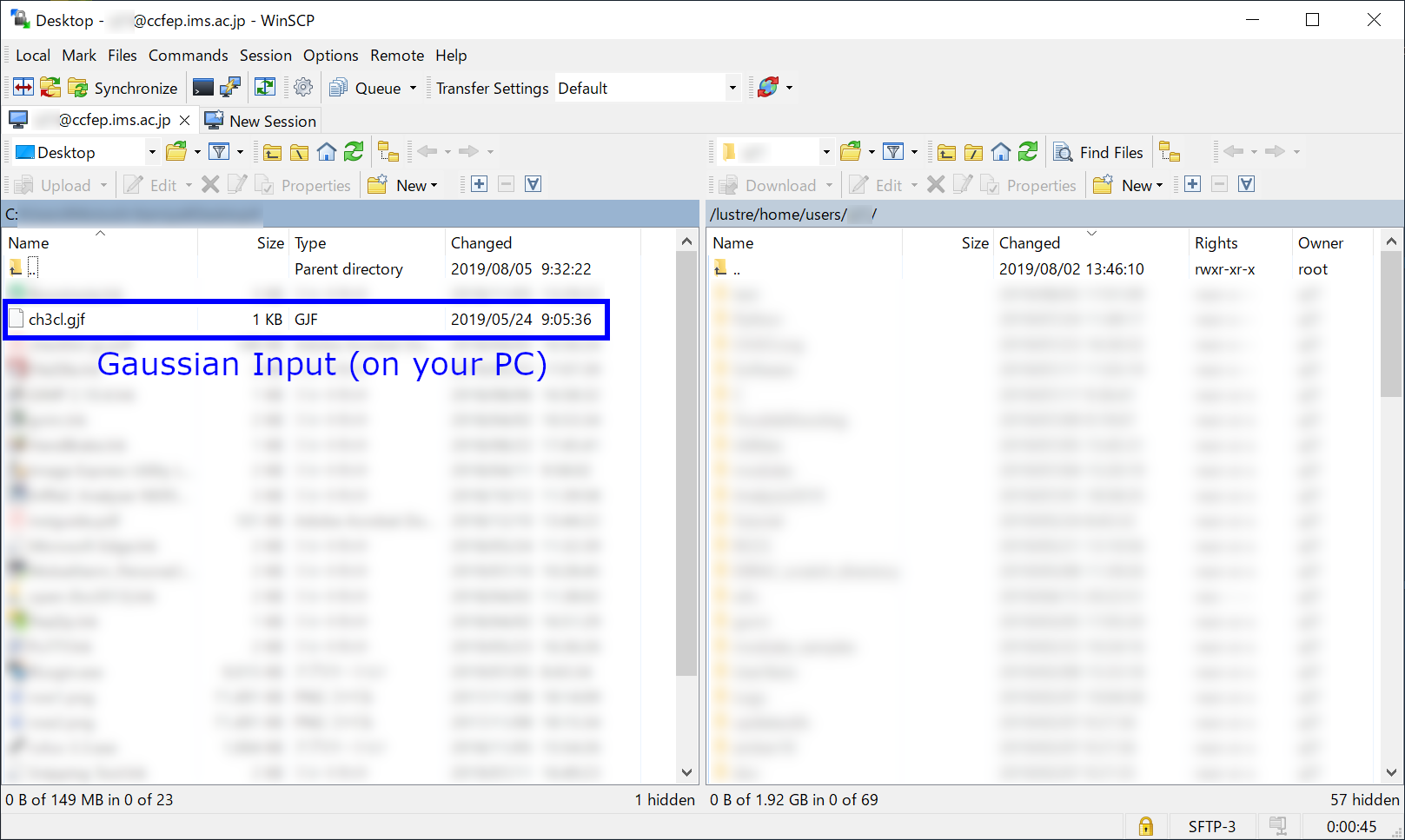
Connect to ccfep.ims.ac.jp, and move to the folder (your PC side) where the Gaussian input file exists. (Note: on RCCS system, /lustre/home/users/(username) and /home/users/(username) are the same path.)
File Transfer (2)
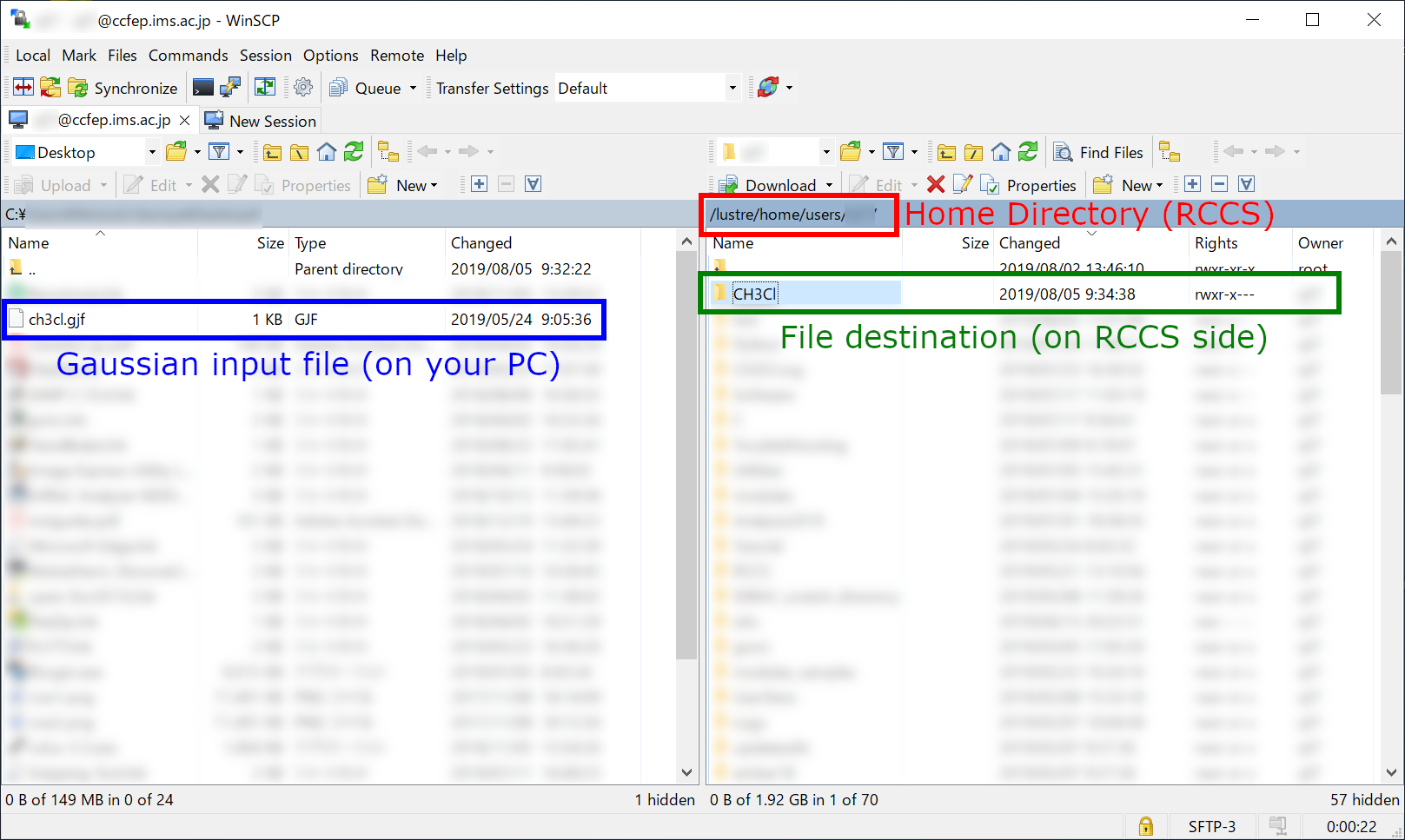
You can create a directory on RCCS side for this job. In this example, we created CH3Cl directory under the home directory (/home/users/(username)/CH3Cl).
File Transfer (3)
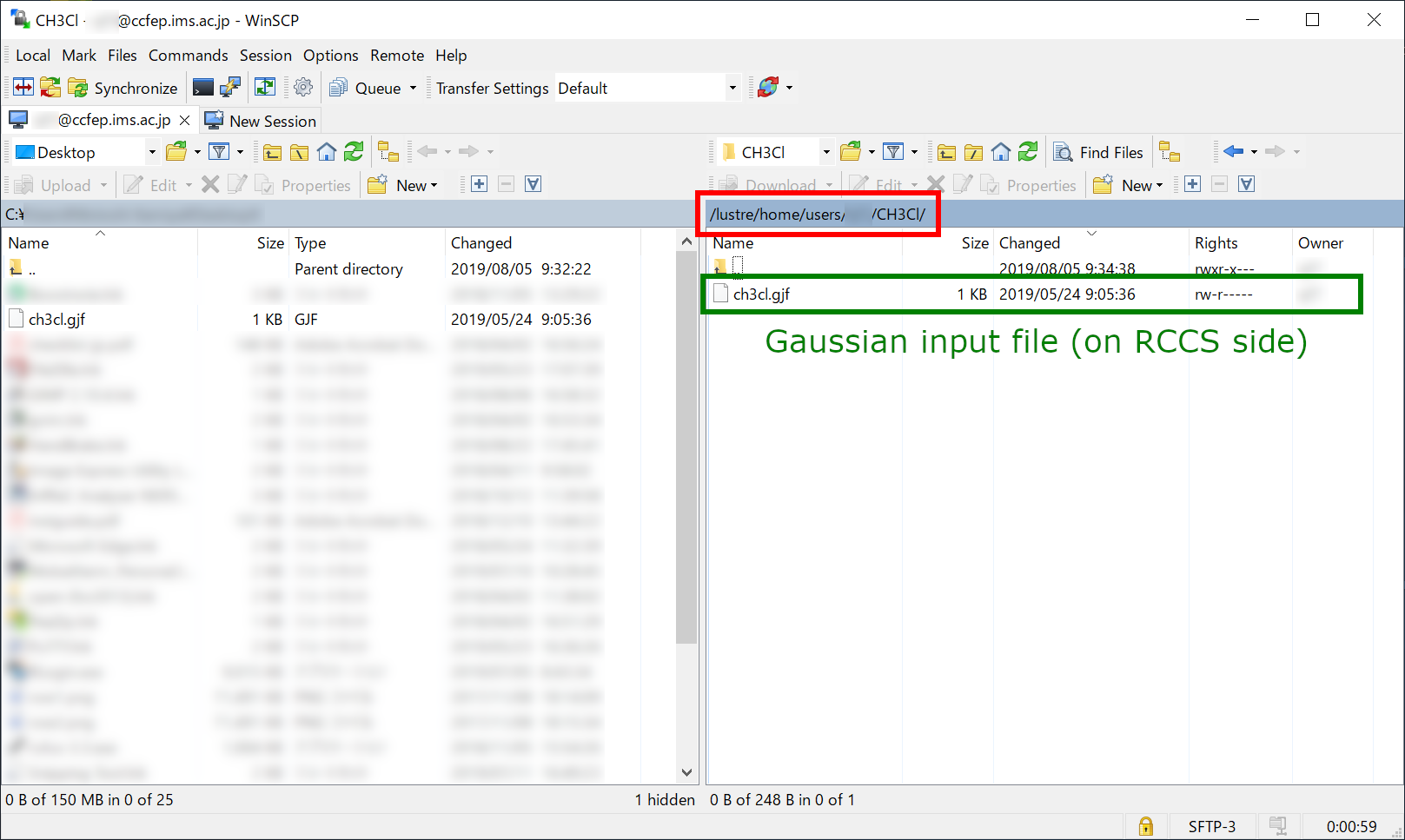
Send input file (e.g. ch3cl.gjf) to RCCS. Once the file transfer finished, you then need to login to RCCS using SSH.
Submit a Gaussian job with g16sub
Login to ccfep.ims.ac.jp (using PuTTY or something). Then, go to the directory where you placed input file (CH3Cl directory in this example) using "cd" command. You can see file list of directories using "ls" command.
Last login: Fri Jan 27 11:24:15 2023 from ***.***.***.***
[user@ccfep3 ~]$ ls CH3Cl/
ch3cl.gjf
[user@ccfep3 ~]$ cd CH3Cl/
[user@ccfep3 CH3Cl]$ ls
ch3cl.gjf
[user@ccfep3 CH3Cl]$
In the target directory (CH3Cl here), submit a Gaussian job with g16sub command.
[user@ccfep3 CH3Cl]$ g16sub ch3cl.gjf
If you submit a job without any arguments other than input file name like above, this job will use 8 CPU cores on a vnode of "core" jobtype. Please note that %mem, %nprocshared, and %cpu in the Gaussian input file will be overwritten by g16sub. The walltime limitation of the job is set to 72 hours. If the job won't finish within the time, the job will be killed by the system.
Some lengthy information like below will be shown when you run the command.
- Basic parameters of the job queue, which involve memory amount per CPU core for example (line began with H( ap) in the box below).
- Summary of file names of your job (colored blue in the box below).
- At the last line, you may see (number).ccpbs1 if your job is successfully submitted. The number (colored red in the box below) is the unique job ID of your job.
[user@ccfep3 CH3Cl]$ g16sub ch3cl.gjf
QUEUE detail
------------------------------------------------------------------------------
QUEUE(MACH) Jobtype MaxMem DefMem TimLim DefCPUs(Min-Max)
------------------------------------------------------------------------------
H( ap) 1.8GB 1.2GB 72:00:00 8(1-128)
------------------------------------------------------------------------------
JOB detail
======================================================================
MOL name(s) : ch3cl
INP file(s) : ch3cl.gjf.ap
OUT file(s) : ch3cl.out
Current dir : /lustre/home/users/***/CH3Cl
SCRATCH dir : /lwork/users/${USER}/${PBS_JOBID}/gaussianQUEUE : H
Memory : 9.6GB
Time limit : 72:00:00
Job script : /lustre/home/users/***/CH3Cl/H-1571896.sh
Input modified : y
======================================================================/usr/local/bin/jsub –q H /lustre/home/users/***/CH3Cl/H-1571896.sh
4008669.ccpbs1
[user@ccfep3 CH3Cl]$
Verify job status
You can see the status of the job with "jobinfo" command. (Job might not be shown immediately after the job submission. Please wait a while in this case.)
[user@ccfep3 CH3Cl]$ jobinfo –c
--------------------------------------------------------------------------------
Queue Job ID Name Status CPUs User/Grp Elaps Node/(Reason)
--------------------------------------------------------------------------------
H 4008669 H-1571896.sh Run 8 user/--- 00:00:00 ccc001
--------------------------------------------------------------------------------
[user@ccfep3 CH3Cl]$
- Job ID (2nd column): the unique ID of your job, corresponding to the last line of g16sub output.
- Status of the job (4th column): "Run" => job is running, "Queue" => job is not yet running.
- Elapsed time and running node name (rightmost two columns): The elapsed time is running/waiting time for "Run"/"Queue" state job, respectively.
- case "Run": the rightmost column shows the node name(s) where your job is running.
- case "Queue": the rightmost column shows the reason why the job is not running.
- (cpu) not enough CPU cores available.
- (long) job walltime is too long (jobs must be finished before the next maintenance). It will run after the next maintenance.
- (other) other reasons. Just submitted jobs will show this reason, too.
Once the job is successfully accepted by the queue, the job exists (and will run eventually) even after you closed the SSH connection.
Job finished...
Once the job finished, you may see new files in the directory. Among them, H_*** (H-1571896.sh, H-1571896.sh.e4008669, H-1571896.sh.o4008669) and ***.ap (ch3cl.gjf.ap) are created by g16sub/g09sub and are not necessary if the job finished successfully.
$ ls
H-1571896.sh H-1571896.sh.o4008669 ch3cl.gjf ch3cl.out
H-1571896.sh.e4008669 ch3cl.chk ch3cl.gjf.ap
$
Gaussian output file (ch3cl.out) can be checked during the job execution using "less" or "tail" command. Checkpoint file (if requested) will also be found in the same directory.
Run formchk
You can run formchk at RCCS login server. But you need to run the following command before running formchk.
In case your login shell is /bin/bash or /bin/zsh:
$ source /apl/gaussian/16c02/g16/bsd/g16.profile
In case your login shell is /bin/csh:
$ source /apl/gaussian/16c02/g16/bsd/g16.login
This command won't output anything. But the setting is loaded and thus you can run formchk now. (You can ignore "PYTHONPATH: Undefined variable." error message. Setting is correctly loaded and formchk is available.)
$ formchk ch3cl.chk
Read checkpoint file ch3cl.chk type Unk
Write formatted file ch3cl.fchk
FChkPn: Coordinates translated and rotated
FChkPn: Coordintesmatch /B/ after translation and rotation
$
If you are not sure what your login shell is, please run "echo $SHELL" command like below.
$ echo $SHELL